import traceback
import warnings
import matplotlib.pyplot as plt
import pandas as pd
from scmdata import ScmRun, run_append
/tmp/ipykernel_782/1318481154.py:5: DeprecationWarning:
Pyarrow will become a required dependency of pandas in the next major release of pandas (pandas 3.0),
(to allow more performant data types, such as the Arrow string type, and better interoperability with other libraries)
but was not found to be installed on your system.
If this would cause problems for you,
please provide us feedback at https://github.com/pandas-dev/pandas/issues/54466
import pandas as pd
/home/docs/checkouts/readthedocs.org/user_builds/scmdata/checkouts/stable/src/scmdata/database/_database.py:9: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
import tqdm.autonotebook as tqdman
Operations
scmdata has limited support for operations with ScmRun instances. Here we provide examples of
how to use them.
Available operations
At present, the following options are available:
add
subtract
divide
multiply
integrate (sum and trapizoidal)
change per unit time (numerical differentiation essentially)
calculate linear regression
shift median of an ensemble
These operations are unit aware so are fairly powerful.
Load some data
We first load some test data.
db_emms = ScmRun("rcp26_emissions.csv", lowercase_cols=True)
db_emms.tail()
| time | 1765-01-01 00:00:00 | 1766-01-01 00:00:00 | 1767-01-01 00:00:00 | 1768-01-01 00:00:00 | 1769-01-01 00:00:00 | 1770-01-01 00:00:00 | 1771-01-01 00:00:00 | 1772-01-01 00:00:00 | 1773-01-01 00:00:00 | 1774-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
| IMAGE | World | RCP26 | Mt NMVOC / yr | Emissions|NMVOC | 0.0 | 1.596875 | 2.292316 | 2.988648 | 3.685897 | 4.384091 | 5.083260 | 5.783435 | 6.484645 | 7.186924 | ... | 125.5104 | 125.5104 | 125.5104 | 125.5104 | 125.5104 | 125.5104 | 125.5104 | 125.5104 | 125.5104 | 125.5104 |
| Mt N / yr | Emissions|NOx | 0.0 | 0.109502 | 0.168038 | 0.226625 | 0.285264 | 0.343956 | 0.402704 | 0.461508 | 0.520371 | 0.579294 | ... | 15.9895 | 15.9895 | 15.9895 | 15.9895 | 15.9895 | 15.9895 | 15.9895 | 15.9895 | 15.9895 | 15.9895 | |||
| Mt OC / yr | Emissions|OC | 0.0 | 0.565920 | 0.781468 | 0.997361 | 1.213611 | 1.430229 | 1.647226 | 1.864613 | 2.082403 | 2.300608 | ... | 25.3311 | 25.3311 | 25.3311 | 25.3311 | 25.3311 | 25.3311 | 25.3311 | 25.3311 | 25.3311 | 25.3311 | |||
| kt SF6 / yr | Emissions|SF6 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.0442 | 0.0442 | 0.0442 | 0.0442 | 0.0442 | 0.0442 | 0.0442 | 0.0442 | 0.0442 | 0.0442 | |||
| Mt S / yr | Emissions|SOx | 0.0 | 0.098883 | 0.116306 | 0.133811 | 0.151398 | 0.169070 | 0.186831 | 0.204683 | 0.222628 | 0.240670 | ... | 6.4552 | 6.4552 | 6.4552 | 6.4552 | 6.4552 | 6.4552 | 6.4552 | 6.4552 | 6.4552 | 6.4552 |
5 rows × 736 columns
db_forcing = ScmRun(
"rcmip-radiative-forcing-annual-means-v4-0-0.csv", lowercase_cols=True
).drop_meta(["mip_era", "activity_id"], inplace=False)
db_forcing.head()
| time | 1750-01-01 00:00:00 | 1751-01-01 00:00:00 | 1752-01-01 00:00:00 | 1753-01-01 00:00:00 | 1754-01-01 00:00:00 | 1755-01-01 00:00:00 | 1756-01-01 00:00:00 | 1757-01-01 00:00:00 | 1758-01-01 00:00:00 | 1759-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
| AIM | World | rcp60 | W/m^2 | Radiative Forcing | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 5.993838 | 5.993838 | 5.993838 | 5.993838 | 5.993838 | 5.993838 | 5.993838 | 5.993838 | 5.993838 | 5.993838 |
| Radiative Forcing|Anthropogenic | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 5.890082 | 5.890082 | 5.890082 | 5.890082 | 5.890082 | 5.890082 | 5.890082 | 5.890082 | 5.890082 | 5.890082 | ||||
| Radiative Forcing|Anthropogenic|Aerosols | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | -0.603691 | -0.603691 | -0.603691 | -0.603691 | -0.603691 | -0.603691 | -0.603691 | -0.603691 | -0.603691 | -0.603691 | ||||
| Radiative Forcing|Anthropogenic|Aerosols|Aerosols-cloud Interactions | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | -0.465404 | -0.465404 | -0.465404 | -0.465404 | -0.465404 | -0.465404 | -0.465404 | -0.465404 | -0.465404 | -0.465404 | ||||
| Radiative Forcing|Anthropogenic|Aerosols|Aerosols-radiation Interactions | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | -0.138286 | -0.138286 | -0.138286 | -0.138286 | -0.138286 | -0.138286 | -0.138286 | -0.138286 | -0.138286 | -0.138286 |
5 rows × 751 columns
Add
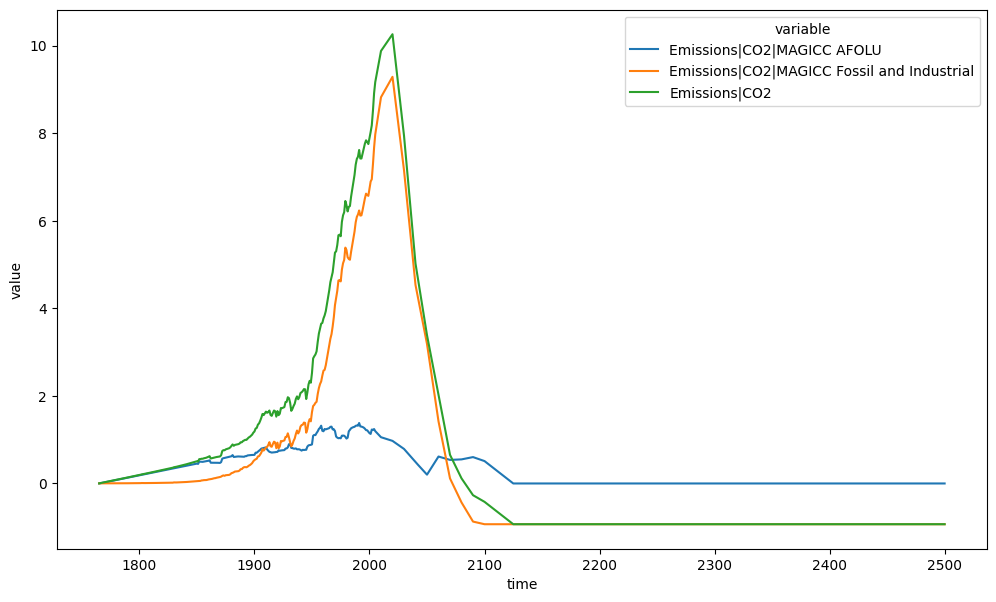
A very simple example is adding two variables together. For example, below we calculate total CO\(_2\) emissions for the RCP2.6 scenario.
emms_co2 = db_emms.filter(variable="Emissions|CO2|MAGICC Fossil and Industrial").add(
db_emms.filter(variable="Emissions|CO2|MAGICC AFOLU"),
op_cols={"variable": "Emissions|CO2"},
)
emms_co2.head()
| time | 1765-01-01 00:00:00 | 1766-01-01 00:00:00 | 1767-01-01 00:00:00 | 1768-01-01 00:00:00 | 1769-01-01 00:00:00 | 1770-01-01 00:00:00 | 1771-01-01 00:00:00 | 1772-01-01 00:00:00 | 1773-01-01 00:00:00 | 1774-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
| IMAGE | World | RCP26 | C * gigametric_ton / a | Emissions|CO2 | 0.003 | 0.008338 | 0.013677 | 0.019015 | 0.024353 | 0.029691 | 0.03603 | 0.041368 | 0.046706 | 0.052045 | ... | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 |
1 rows × 736 columns
ax = plt.figure(figsize=(12, 7)).add_subplot(111)
plt_df = run_append([db_emms, emms_co2])
plt_df.filter(variable="*CO2*").lineplot(hue="variable", ax=ax)
<Axes: xlabel='time', ylabel='value'>
The op_cols argument tells scmdata which columns to ignore when aligning the data
and what value to give this column in the output. So we could do the same calculation
but give the output a different name as shown below.
emms_co2_different_name = db_emms.filter(
variable="Emissions|CO2|MAGICC Fossil and Industrial"
).add(
db_emms.filter(variable="Emissions|CO2|MAGICC AFOLU"),
op_cols={"variable": "Emissions|CO2|Total"},
)
emms_co2_different_name.head()
| time | 1765-01-01 00:00:00 | 1766-01-01 00:00:00 | 1767-01-01 00:00:00 | 1768-01-01 00:00:00 | 1769-01-01 00:00:00 | 1770-01-01 00:00:00 | 1771-01-01 00:00:00 | 1772-01-01 00:00:00 | 1773-01-01 00:00:00 | 1774-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
| IMAGE | World | RCP26 | C * gigametric_ton / a | Emissions|CO2|Total | 0.003 | 0.008338 | 0.013677 | 0.019015 | 0.024353 | 0.029691 | 0.03603 | 0.041368 | 0.046706 | 0.052045 | ... | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 | -0.9308 |
1 rows × 736 columns
Subtract
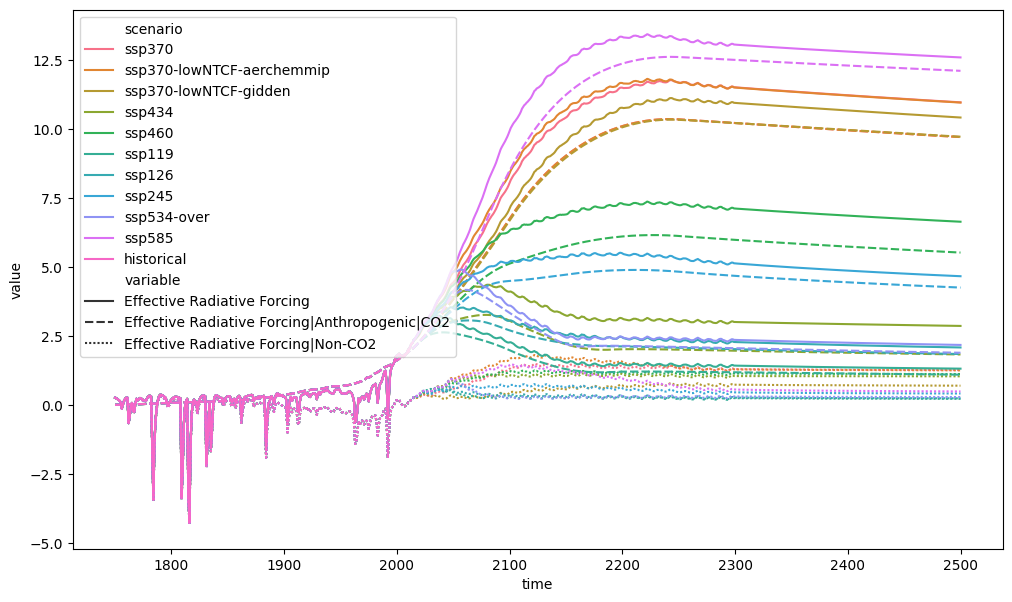
Subtraction works much the same way. Below we calculate the total effective radiative forcing and CO\(_2\) effective radiative forcing in the RCMIP data.
non_co2_rf = db_forcing.filter(variable="Effective Radiative Forcing").subtract(
db_forcing.filter(variable="Effective Radiative Forcing|Anthropogenic|CO2"),
op_cols={"variable": "Effective Radiative Forcing|Non-CO2"},
)
non_co2_rf.head()
| time | 1750-01-01 00:00:00 | 1751-01-01 00:00:00 | 1752-01-01 00:00:00 | 1753-01-01 00:00:00 | 1754-01-01 00:00:00 | 1755-01-01 00:00:00 | 1756-01-01 00:00:00 | 1757-01-01 00:00:00 | 1758-01-01 00:00:00 | 1759-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
| AIM/CGE | World | ssp370 | watt / meter ** 2 | Effective Radiative Forcing|Non-CO2 | 0.259367 | 0.241965 | 0.213009 | 0.177158 | 0.142201 | 0.094554 | -0.137892 | 0.010089 | 0.140326 | 0.218701 | ... | 1.259420 | 1.259326 | 1.259229 | 1.259130 | 1.259034 | 1.258938 | 1.258848 | 1.258752 | 1.258657 | 1.258609 |
| ssp370-lowNTCF-aerchemmip | watt / meter ** 2 | Effective Radiative Forcing|Non-CO2 | 0.259367 | 0.241965 | 0.213009 | 0.177158 | 0.142201 | 0.094554 | -0.137892 | 0.010089 | 0.140326 | 0.218701 | ... | 1.250176 | 1.250081 | 1.249985 | 1.249885 | 1.249789 | 1.249693 | 1.249603 | 1.249508 | 1.249412 | 1.249365 | ||
| ssp370-lowNTCF-gidden | watt / meter ** 2 | Effective Radiative Forcing|Non-CO2 | 0.259367 | 0.241965 | 0.213009 | 0.177158 | 0.142201 | 0.094554 | -0.137892 | 0.010089 | 0.140326 | 0.218701 | ... | 0.695533 | 0.695413 | 0.695297 | 0.695182 | 0.695067 | 0.694952 | 0.694830 | 0.694715 | 0.694600 | 0.694547 | ||
| GCAM4 | World | ssp434 | watt / meter ** 2 | Effective Radiative Forcing|Non-CO2 | 0.259367 | 0.241965 | 0.213009 | 0.177158 | 0.142201 | 0.094554 | -0.137892 | 0.010089 | 0.140326 | 0.218701 | ... | 1.033839 | 1.033813 | 1.033785 | 1.033760 | 1.033735 | 1.033710 | 1.033683 | 1.033658 | 1.033626 | 1.033616 |
| ssp460 | watt / meter ** 2 | Effective Radiative Forcing|Non-CO2 | 0.259367 | 0.241965 | 0.213009 | 0.177158 | 0.142201 | 0.094554 | -0.137892 | 0.010089 | 0.140326 | 0.218701 | ... | 1.116804 | 1.116756 | 1.116709 | 1.116659 | 1.116612 | 1.116565 | 1.116519 | 1.116472 | 1.116429 | 1.116403 |
5 rows × 751 columns
ax = plt.figure(figsize=(12, 7)).add_subplot(111)
plt_df_forcing = run_append([db_forcing, non_co2_rf])
plt_df_forcing.filter(
variable=["Effective Radiative Forcing", "Effective*CO2*"]
).lineplot(style="variable")
<Axes: xlabel='time', ylabel='value'>
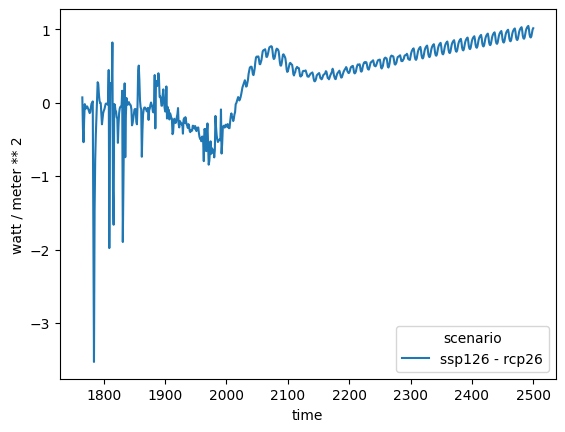
We could also calculate the difference between some SSP and RCP scenarios. The first thing to try would be to simply subtract the SSP126 total effective radiative forcing from the RCP26 total radiative forcing.
try:
ssp126_minus_rcp26 = db_forcing.filter(
scenario="ssp126", variable="Effective Radiative Forcing"
).subtract(
db_forcing.filter(scenario="rcp26", variable="Radiative Forcing"),
op_cols={
"scenario": "ssp126 - rcp26",
},
)
except KeyError:
traceback.print_exc(limit=0, chain=False)
KeyError: "No equivalent in `other` for [('model', 'IMAGE'), ('region', 'World'), ('variable', 'Effective Radiative Forcing')]"
Doing this gives us a KeyError. The reason is that the SSP126 variable
is Effective Radiative Forcing whilst the RCP26 variable is Radiative Forcing
hence the two datasets don’t align. We can work around this using the op_cols argument.
ssp126_minus_rcp26 = db_forcing.filter(
scenario="ssp126", variable="Effective Radiative Forcing"
).subtract(
db_forcing.filter(scenario="rcp26", variable="Radiative Forcing"),
op_cols={
"scenario": "ssp126 - rcp26",
"variable": "RF",
},
)
ssp126_minus_rcp26.lineplot()
<Axes: xlabel='time', ylabel='watt / meter ** 2'>
We could create plots of all the differences as shown below.
ssp_rcp_diffs = []
for target in ["26", "45", "60", "85"]:
ssp = db_forcing.filter(
scenario=f"ssp*{target}",
variable="Effective Radiative Forcing",
)
ssp_scen = ssp.get_unique_meta("scenario", no_duplicates=True)
ssp_model = ssp.get_unique_meta("model", no_duplicates=True)
rcp = db_forcing.filter(scenario=f"rcp{target}", variable="Radiative Forcing")
rcp_scen = rcp.get_unique_meta("scenario", no_duplicates=True)
rcp_model = rcp.get_unique_meta("model", no_duplicates=True)
ssp_rcp_diff = ssp.subtract(
rcp,
op_cols={
"scenario": f"{ssp_scen} - {rcp_scen}",
"model": f"{ssp_model} - {rcp_model}",
"variable": "RF",
},
)
ssp_rcp_diffs.append(ssp_rcp_diff)
ssp_rcp_diffs = run_append(ssp_rcp_diffs)
ssp_rcp_diffs.head()
| time | 1750-01-01 00:00:00 | 1751-01-01 00:00:00 | 1752-01-01 00:00:00 | 1753-01-01 00:00:00 | 1754-01-01 00:00:00 | 1755-01-01 00:00:00 | 1756-01-01 00:00:00 | 1757-01-01 00:00:00 | 1758-01-01 00:00:00 | 1759-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
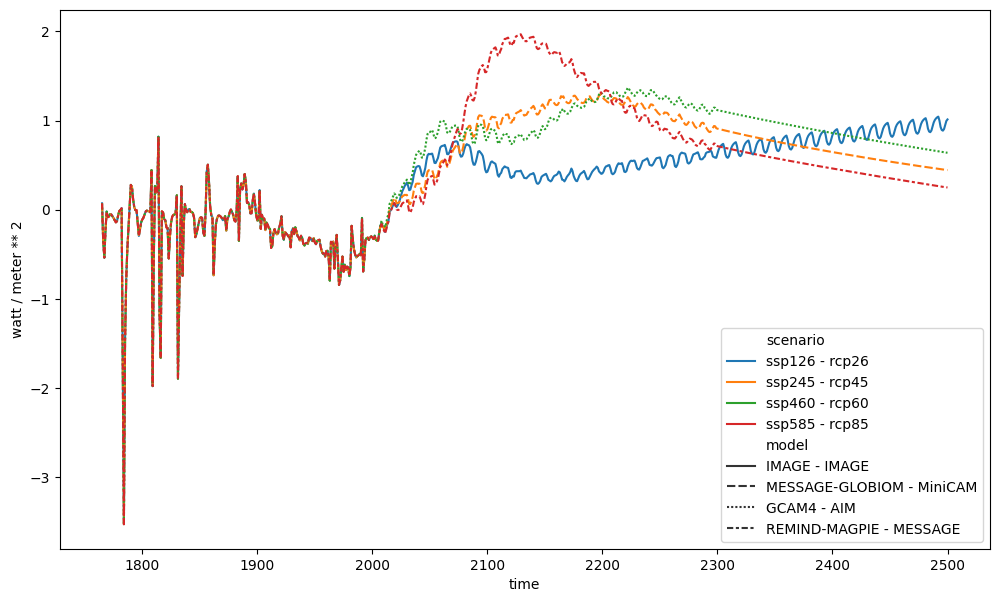
| IMAGE - IMAGE | World | ssp126 - rcp26 | watt / meter ** 2 | RF | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 1.036063 | 1.044192 | 0.998926 | 0.927014 | 0.894074 | 0.886604 | 0.904720 | 0.950169 | 0.995562 | 1.012583 |
| MESSAGE-GLOBIOM - MiniCAM | World | ssp245 - rcp45 | watt / meter ** 2 | RF | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.460000 | 0.458162 | 0.456325 | 0.454487 | 0.452659 | 0.450835 | 0.449006 | 0.447191 | 0.445375 | 0.444465 |
| GCAM4 - AIM | World | ssp460 - rcp60 | watt / meter ** 2 | RF | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.656893 | 0.654786 | 0.652687 | 0.650594 | 0.648503 | 0.646411 | 0.644319 | 0.642235 | 0.640155 | 0.639114 |
| REMIND-MAGPIE - MESSAGE | World | ssp585 - rcp85 | watt / meter ** 2 | RF | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 0.266282 | 0.264204 | 0.262160 | 0.260144 | 0.258118 | 0.256072 | 0.254027 | 0.252006 | 0.250012 | 0.248969 |
4 rows × 751 columns
ax = plt.figure(figsize=(12, 7)).add_subplot(111)
ssp_rcp_diffs.lineplot(ax=ax, style="model")
<Axes: xlabel='time', ylabel='watt / meter ** 2'>
Divide
The divide (and multiply) operations clearly have to also be aware of units.
Thanks to Pint’s pandas interface,
this can happen automatically. For example, in our calculation below the units are
automatically returned as dimensionless.
ssp126_to_rcp26 = db_forcing.filter(
scenario="ssp126", variable="Effective Radiative Forcing"
).divide(
db_forcing.filter(scenario="rcp26", variable="Radiative Forcing"),
op_cols={
"scenario": "ssp126 / rcp26",
"variable": "RF",
},
)
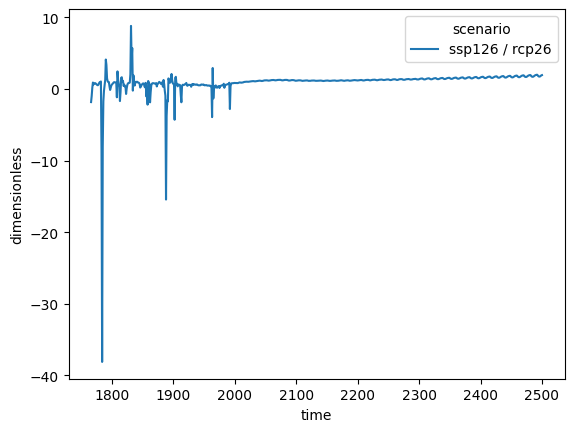
ssp126_to_rcp26.head()
| time | 1750-01-01 00:00:00 | 1751-01-01 00:00:00 | 1752-01-01 00:00:00 | 1753-01-01 00:00:00 | 1754-01-01 00:00:00 | 1755-01-01 00:00:00 | 1756-01-01 00:00:00 | 1757-01-01 00:00:00 | 1758-01-01 00:00:00 | 1759-01-01 00:00:00 | ... | 2491-01-01 00:00:00 | 2492-01-01 00:00:00 | 2493-01-01 00:00:00 | 2494-01-01 00:00:00 | 2495-01-01 00:00:00 | 2496-01-01 00:00:00 | 2497-01-01 00:00:00 | 2498-01-01 00:00:00 | 2499-01-01 00:00:00 | 2500-01-01 00:00:00 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| model | region | scenario | unit | variable | |||||||||||||||||||||
| IMAGE | World | ssp126 / rcp26 | dimensionless | RF | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 1.987642 | 2.003883 | 1.920958 | 1.802047 | 1.752583 | 1.742102 | 1.769413 | 1.841106 | 1.918814 | 1.949772 |
1 rows × 751 columns
ssp126_to_rcp26.lineplot()
<Axes: xlabel='time', ylabel='dimensionless'>
Integrate
We can also integrate our data. As previously, thanks to Pint and some other work, this operation is also unit aware.
There are two methods of integration available, cumtrapz and cumsum.
The method that should be used depends on the data you are integrating,
specifically whether the data are piecewise linear or piecewise constant.
Annual timeseries of emissions are piecewise constant (each value represents
a total which is constant over an year) so should use the cumsum method.
Other output such as effective radiative forcing, concentrations or decadal
timeseries of emissions, represent a point estimate or an average over a period.
These timeseries are therefore piecewise linear and should use the cumtrapz
method.
with warnings.catch_warnings():
# Ignore warning about nans in the historical timeseries
warnings.filterwarnings("ignore", module="scmdata.ops")
# Radiative Forcings are piecewise-linear so `cumtrapz` should be used
erf_integral = (
db_forcing.filter(variable="Effective Radiative Forcing")
.cumtrapz()
.convert_unit("TJ / m^2")
)
erf_integral
<ScmRun (timeseries: 11, timepoints: 751)>
Time:
Start: 1750-01-01T00:00:00
End: 2500-01-01T00:00:00
Meta:
model region scenario unit \
0 AIM/CGE World ssp370 TJ / m^2
1 AIM/CGE World ssp370-lowNTCF-aerchemmip TJ / m^2
2 AIM/CGE World ssp370-lowNTCF-gidden TJ / m^2
3 GCAM4 World ssp434 TJ / m^2
4 GCAM4 World ssp460 TJ / m^2
5 IMAGE World ssp119 TJ / m^2
6 IMAGE World ssp126 TJ / m^2
7 MESSAGE-GLOBIOM World ssp245 TJ / m^2
8 REMIND-MAGPIE World ssp534-over TJ / m^2
9 REMIND-MAGPIE World ssp585 TJ / m^2
10 unspecified World historical TJ / m^2
variable
0 Cumulative Effective Radiative Forcing
1 Cumulative Effective Radiative Forcing
2 Cumulative Effective Radiative Forcing
3 Cumulative Effective Radiative Forcing
4 Cumulative Effective Radiative Forcing
5 Cumulative Effective Radiative Forcing
6 Cumulative Effective Radiative Forcing
7 Cumulative Effective Radiative Forcing
8 Cumulative Effective Radiative Forcing
9 Cumulative Effective Radiative Forcing
10 Cumulative Effective Radiative Forcing
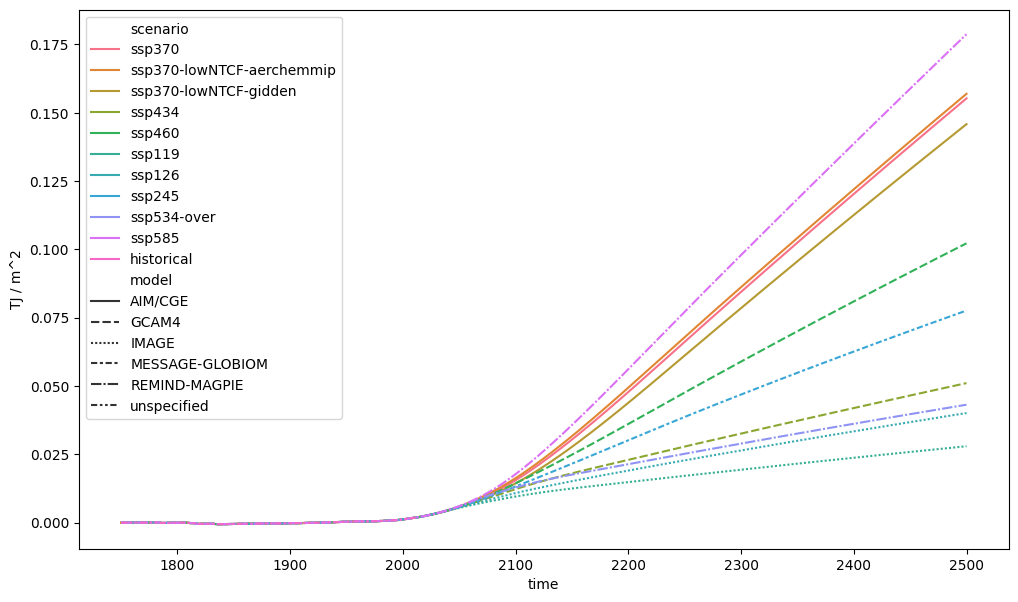
ax = plt.figure(figsize=(12, 7)).add_subplot(111)
erf_integral.lineplot(ax=ax, style="model")
<Axes: xlabel='time', ylabel='TJ / m^2'>
with warnings.catch_warnings():
# Ignore warning about nans in the historical timeseries
warnings.filterwarnings("ignore", module="scmdata.ops")
# Emissions are piecewise-constant so `cumsum` should be used
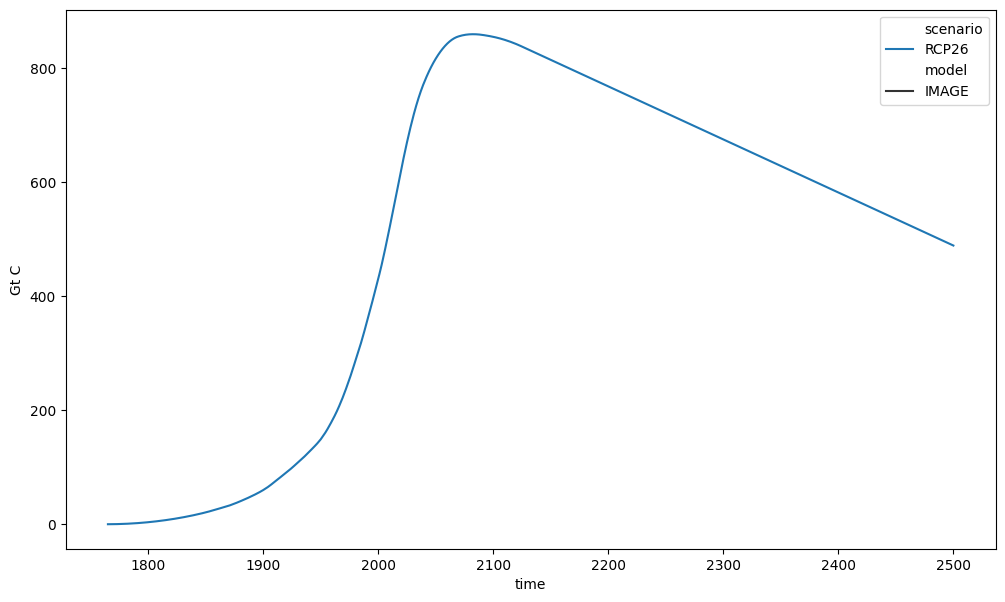
cumulative_emissions = emms_co2.cumsum().convert_unit("Gt C")
cumulative_emissions
<ScmRun (timeseries: 1, timepoints: 736)>
Time:
Start: 1765-01-01T00:00:00
End: 2500-01-01T00:00:00
Meta:
model region scenario unit variable
0 IMAGE World RCP26 Gt C Cumulative Emissions|CO2
ax = plt.figure(figsize=(12, 7)).add_subplot(111)
cumulative_emissions.lineplot(ax=ax, style="model")
<Axes: xlabel='time', ylabel='Gt C'>
Time deltas
We can also calculate the change per unit time of our data. As previously, thanks to Pint and some other work, this operation is also unit aware.
with warnings.catch_warnings():
# Ignore warning about nans in the historical timeseries
warnings.filterwarnings("ignore", module="scmdata.ops")
erf_delta = (
db_forcing.filter(variable="Effective Radiative Forcing")
.delta_per_delta_time()
.convert_unit("W / m^2 / yr")
)
erf_delta
<ScmRun (timeseries: 11, timepoints: 750)>
Time:
Start: 1750-07-02T12:00:00
End: 2499-07-02T12:00:00
Meta:
model region scenario unit \
0 AIM/CGE World ssp370 W / m^2 / yr
1 AIM/CGE World ssp370-lowNTCF-aerchemmip W / m^2 / yr
2 AIM/CGE World ssp370-lowNTCF-gidden W / m^2 / yr
3 GCAM4 World ssp434 W / m^2 / yr
4 GCAM4 World ssp460 W / m^2 / yr
5 IMAGE World ssp119 W / m^2 / yr
6 IMAGE World ssp126 W / m^2 / yr
7 MESSAGE-GLOBIOM World ssp245 W / m^2 / yr
8 REMIND-MAGPIE World ssp534-over W / m^2 / yr
9 REMIND-MAGPIE World ssp585 W / m^2 / yr
10 unspecified World historical W / m^2 / yr
variable
0 Delta Effective Radiative Forcing
1 Delta Effective Radiative Forcing
2 Delta Effective Radiative Forcing
3 Delta Effective Radiative Forcing
4 Delta Effective Radiative Forcing
5 Delta Effective Radiative Forcing
6 Delta Effective Radiative Forcing
7 Delta Effective Radiative Forcing
8 Delta Effective Radiative Forcing
9 Delta Effective Radiative Forcing
10 Delta Effective Radiative Forcing
fig, axes = plt.subplots(figsize=(16, 9), ncols=2)
erf_delta.lineplot(ax=axes[0], style="model")
erf_delta.filter(year=range(2000, 2500)).lineplot(
ax=axes[1], style="model", legend=False
)
axes[1].set_ylim([-0.1, 0.2])
(-0.1, 0.2)
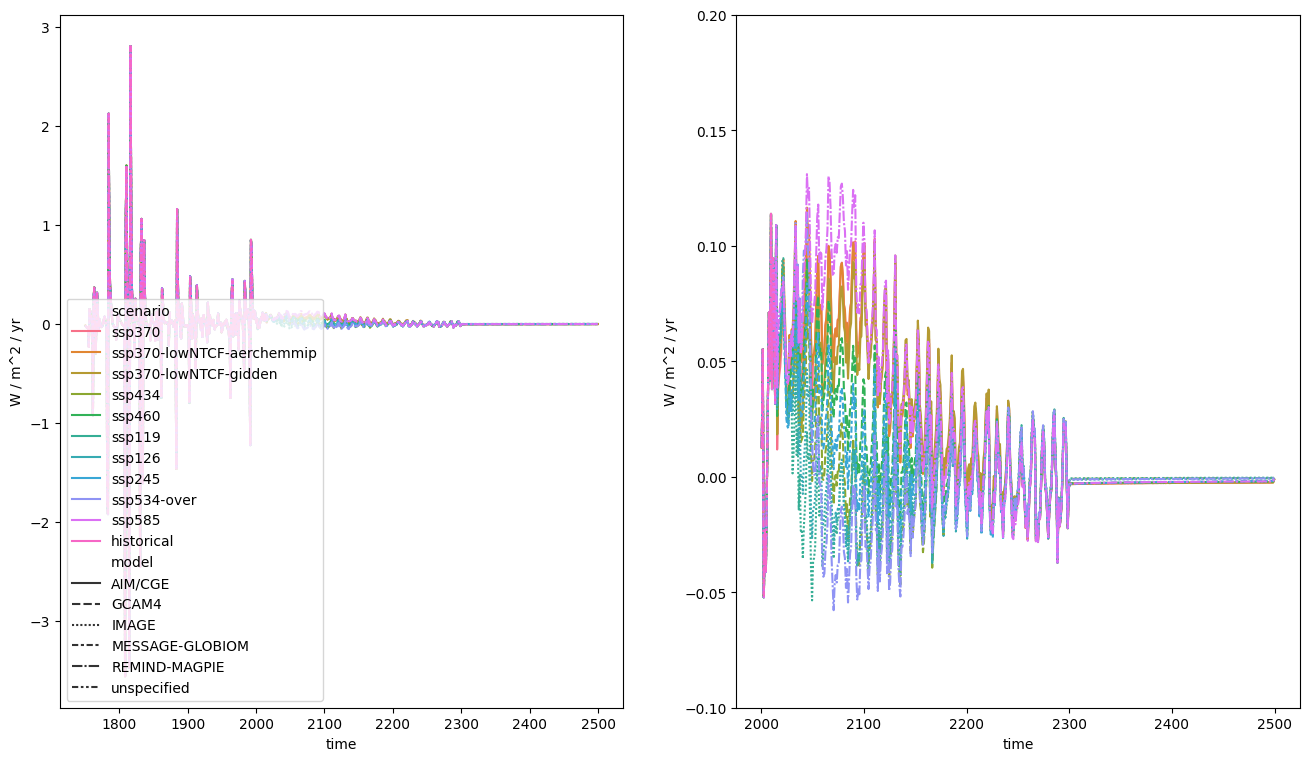
Regression
We can also calculate linear regressions of our data. As previously, thanks to Pint and some other work, this operation is also unit aware.
erf_total = db_forcing.filter(variable="Effective Radiative Forcing").filter(
scenario="historical", keep=False
)
erf_total_for_reg = erf_total.filter(year=range(2010, 2050))
The default return type of linear_regression is a list of dictionaries.
linear_regression_raw = erf_total_for_reg.linear_regression()
type(linear_regression_raw)
list
type(linear_regression_raw[0])
dict
If we want, we can make a DataFrame from this list.
linear_regression_df = pd.DataFrame(linear_regression_raw)
linear_regression_df
| model | region | scenario | variable | gradient | intercept | |
|---|---|---|---|---|---|---|
| 0 | AIM/CGE | World | ssp370 | Effective Radiative Forcing | 1.90373454366716e-09 watt / meter ** 2 / second | -0.42795992093158064 watt / meter ** 2 |
| 1 | AIM/CGE | World | ssp370-lowNTCF-aerchemmip | Effective Radiative Forcing | 2.196587535316519e-09 watt / meter ** 2 / second | -0.8448632244177651 watt / meter ** 2 |
| 2 | AIM/CGE | World | ssp370-lowNTCF-gidden | Effective Radiative Forcing | 1.599468808895454e-09 watt / meter ** 2 / second | 0.057473556012937535 watt / meter ** 2 |
| 3 | GCAM4 | World | ssp434 | Effective Radiative Forcing | 1.513761825121089e-09 watt / meter ** 2 / second | 0.21738181303589565 watt / meter ** 2 |
| 4 | GCAM4 | World | ssp460 | Effective Radiative Forcing | 1.8242980920680464e-09 watt / meter ** 2 / second | -0.2804321989242344 watt / meter ** 2 |
| 5 | IMAGE | World | ssp119 | Effective Radiative Forcing | 8.935038882642927e-10 watt / meter ** 2 / second | 1.2017704356912693 watt / meter ** 2 |
| 6 | IMAGE | World | ssp126 | Effective Radiative Forcing | 1.2279126589716315e-09 watt / meter ** 2 / second | 0.6544862441567445 watt / meter ** 2 |
| 7 | MESSAGE-GLOBIOM | World | ssp245 | Effective Radiative Forcing | 1.6273999739537484e-09 watt / meter ** 2 / second | 0.033991206478731474 watt / meter ** 2 |
| 8 | REMIND-MAGPIE | World | ssp534-over | Effective Radiative Forcing | 2.2703911252022266e-09 watt / meter ** 2 / second | -0.9204663925813401 watt / meter ** 2 |
| 9 | REMIND-MAGPIE | World | ssp585 | Effective Radiative Forcing | 2.2626366011973838e-09 watt / meter ** 2 / second | -0.9168810372684146 watt / meter ** 2 |
Alternately, we can request only the gradients or only the intercepts (noting that intercepts are calculated using a time base which has zero at 1970-01-01 again).
erf_total_for_reg.linear_regression_gradient("W / m^2 / yr")
| model | region | scenario | variable | gradient | unit | |
|---|---|---|---|---|---|---|
| 0 | AIM/CGE | World | ssp370 | Effective Radiative Forcing | 0.060077 | W / m^2 / yr |
| 1 | AIM/CGE | World | ssp370-lowNTCF-aerchemmip | Effective Radiative Forcing | 0.069319 | W / m^2 / yr |
| 2 | AIM/CGE | World | ssp370-lowNTCF-gidden | Effective Radiative Forcing | 0.050475 | W / m^2 / yr |
| 3 | GCAM4 | World | ssp434 | Effective Radiative Forcing | 0.047771 | W / m^2 / yr |
| 4 | GCAM4 | World | ssp460 | Effective Radiative Forcing | 0.057570 | W / m^2 / yr |
| 5 | IMAGE | World | ssp119 | Effective Radiative Forcing | 0.028197 | W / m^2 / yr |
| 6 | IMAGE | World | ssp126 | Effective Radiative Forcing | 0.038750 | W / m^2 / yr |
| 7 | MESSAGE-GLOBIOM | World | ssp245 | Effective Radiative Forcing | 0.051357 | W / m^2 / yr |
| 8 | REMIND-MAGPIE | World | ssp534-over | Effective Radiative Forcing | 0.071648 | W / m^2 / yr |
| 9 | REMIND-MAGPIE | World | ssp585 | Effective Radiative Forcing | 0.071403 | W / m^2 / yr |
erf_total_for_reg.linear_regression_intercept("W / m^2")
| model | region | scenario | variable | intercept | unit | |
|---|---|---|---|---|---|---|
| 0 | AIM/CGE | World | ssp370 | Effective Radiative Forcing | -0.427960 | W / m^2 |
| 1 | AIM/CGE | World | ssp370-lowNTCF-aerchemmip | Effective Radiative Forcing | -0.844863 | W / m^2 |
| 2 | AIM/CGE | World | ssp370-lowNTCF-gidden | Effective Radiative Forcing | 0.057474 | W / m^2 |
| 3 | GCAM4 | World | ssp434 | Effective Radiative Forcing | 0.217382 | W / m^2 |
| 4 | GCAM4 | World | ssp460 | Effective Radiative Forcing | -0.280432 | W / m^2 |
| 5 | IMAGE | World | ssp119 | Effective Radiative Forcing | 1.201770 | W / m^2 |
| 6 | IMAGE | World | ssp126 | Effective Radiative Forcing | 0.654486 | W / m^2 |
| 7 | MESSAGE-GLOBIOM | World | ssp245 | Effective Radiative Forcing | 0.033991 | W / m^2 |
| 8 | REMIND-MAGPIE | World | ssp534-over | Effective Radiative Forcing | -0.920466 | W / m^2 |
| 9 | REMIND-MAGPIE | World | ssp585 | Effective Radiative Forcing | -0.916881 | W / m^2 |
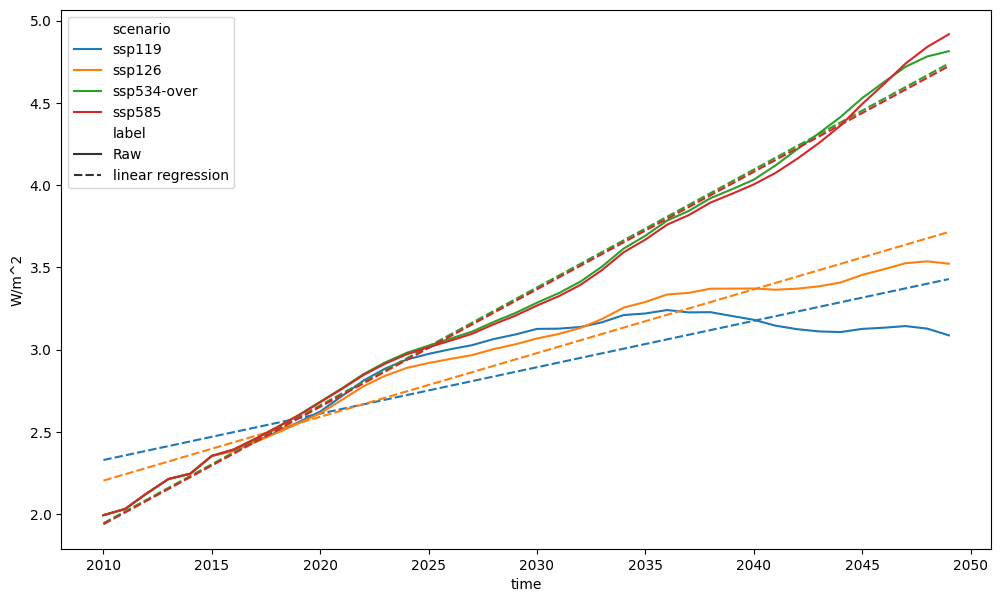
If we want to plot the regressions, we can request an ScmRun instance based
on the linear regressions is returned using the linear_regression_scmrun method.
linear_regression = erf_total_for_reg.linear_regression_scmrun()
linear_regression["label"] = "linear regression"
linear_regression
<ScmRun (timeseries: 10, timepoints: 40)>
Time:
Start: 2010-01-01T00:00:00
End: 2049-01-01T00:00:00
Meta:
label model region scenario \
0 linear regression AIM/CGE World ssp370
1 linear regression AIM/CGE World ssp370-lowNTCF-aerchemmip
2 linear regression AIM/CGE World ssp370-lowNTCF-gidden
3 linear regression GCAM4 World ssp434
4 linear regression GCAM4 World ssp460
5 linear regression IMAGE World ssp119
6 linear regression IMAGE World ssp126
7 linear regression MESSAGE-GLOBIOM World ssp245
8 linear regression REMIND-MAGPIE World ssp534-over
9 linear regression REMIND-MAGPIE World ssp585
unit variable
0 W/m^2 Effective Radiative Forcing
1 W/m^2 Effective Radiative Forcing
2 W/m^2 Effective Radiative Forcing
3 W/m^2 Effective Radiative Forcing
4 W/m^2 Effective Radiative Forcing
5 W/m^2 Effective Radiative Forcing
6 W/m^2 Effective Radiative Forcing
7 W/m^2 Effective Radiative Forcing
8 W/m^2 Effective Radiative Forcing
9 W/m^2 Effective Radiative Forcing
erf_total_for_reg["label"] = "Raw"
pdf = run_append([erf_total_for_reg, linear_regression])
fig, axes = plt.subplots(figsize=(12, 7))
pdf.filter(scenario=["ssp1*", "ssp5*"]).lineplot(ax=axes, style="label")
<Axes: xlabel='time', ylabel='W/m^2'>
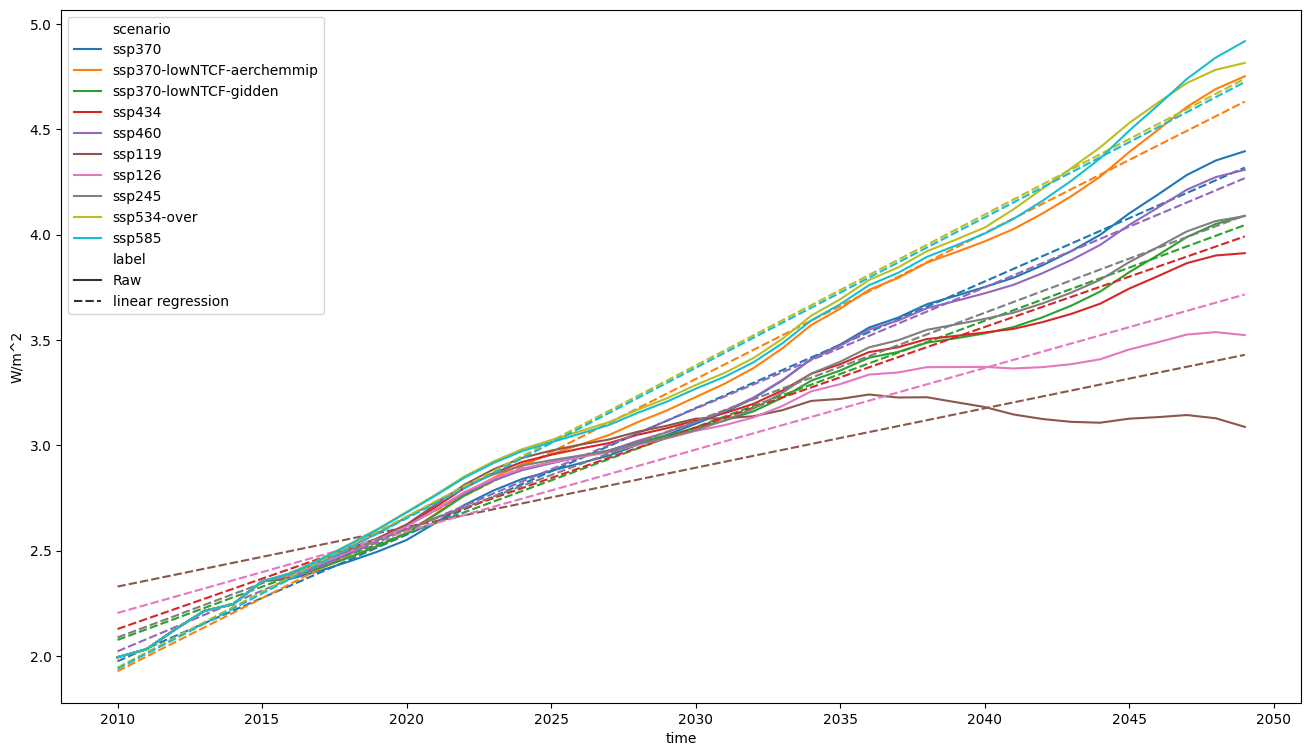
fig, axes = plt.subplots(figsize=(16, 9))
pdf.lineplot(ax=axes, style="label")
<Axes: xlabel='time', ylabel='W/m^2'>
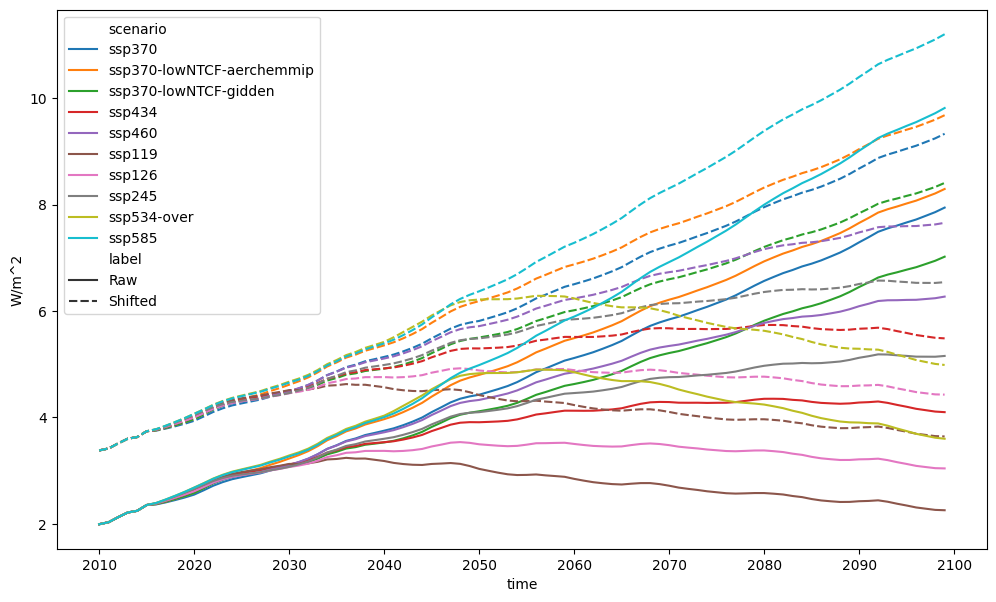
Shift median
Sometimes we wish to simply move an ensemble of timeseries so that its median
matches some value, whilst preserving the spread of the ensemble. This can be
done with the adjust_median_to_target method. For example, let’s say that we
wanted to shift our ensemble of forcing values so that their 2030 median was equal
to 4.5 (god knows why we would want to do this, but it will serve as an example).
erf_total = db_forcing.filter(variable="Effective Radiative Forcing").filter(
scenario="historical", keep=False
)
erf_total_for_shift = erf_total.filter(year=range(2010, 2100))
erf_total_for_shift["label"] = "Raw"
erf_total_for_shift
<ScmRun (timeseries: 10, timepoints: 90)>
Time:
Start: 2010-01-01T00:00:00
End: 2099-01-01T00:00:00
Meta:
label model region scenario unit \
58 Raw AIM/CGE World ssp370 W/m^2
77 Raw AIM/CGE World ssp370-lowNTCF-aerchemmip W/m^2
96 Raw AIM/CGE World ssp370-lowNTCF-gidden W/m^2
115 Raw GCAM4 World ssp434 W/m^2
134 Raw GCAM4 World ssp460 W/m^2
211 Raw IMAGE World ssp119 W/m^2
230 Raw IMAGE World ssp126 W/m^2
307 Raw MESSAGE-GLOBIOM World ssp245 W/m^2
384 Raw REMIND-MAGPIE World ssp534-over W/m^2
403 Raw REMIND-MAGPIE World ssp585 W/m^2
variable
58 Effective Radiative Forcing
77 Effective Radiative Forcing
96 Effective Radiative Forcing
115 Effective Radiative Forcing
134 Effective Radiative Forcing
211 Effective Radiative Forcing
230 Effective Radiative Forcing
307 Effective Radiative Forcing
384 Effective Radiative Forcing
403 Effective Radiative Forcing
erf_total_shifted = erf_total_for_shift.adjust_median_to_target(
target=4.5,
evaluation_period=2030,
process_over=("scenario", "model"),
)
erf_total_shifted["label"] = "Shifted"
pdf = run_append([erf_total_for_shift, erf_total_shifted])
fig, axes = plt.subplots(figsize=(12, 7))
pdf.lineplot(ax=axes, style="label")
<Axes: xlabel='time', ylabel='W/m^2'>
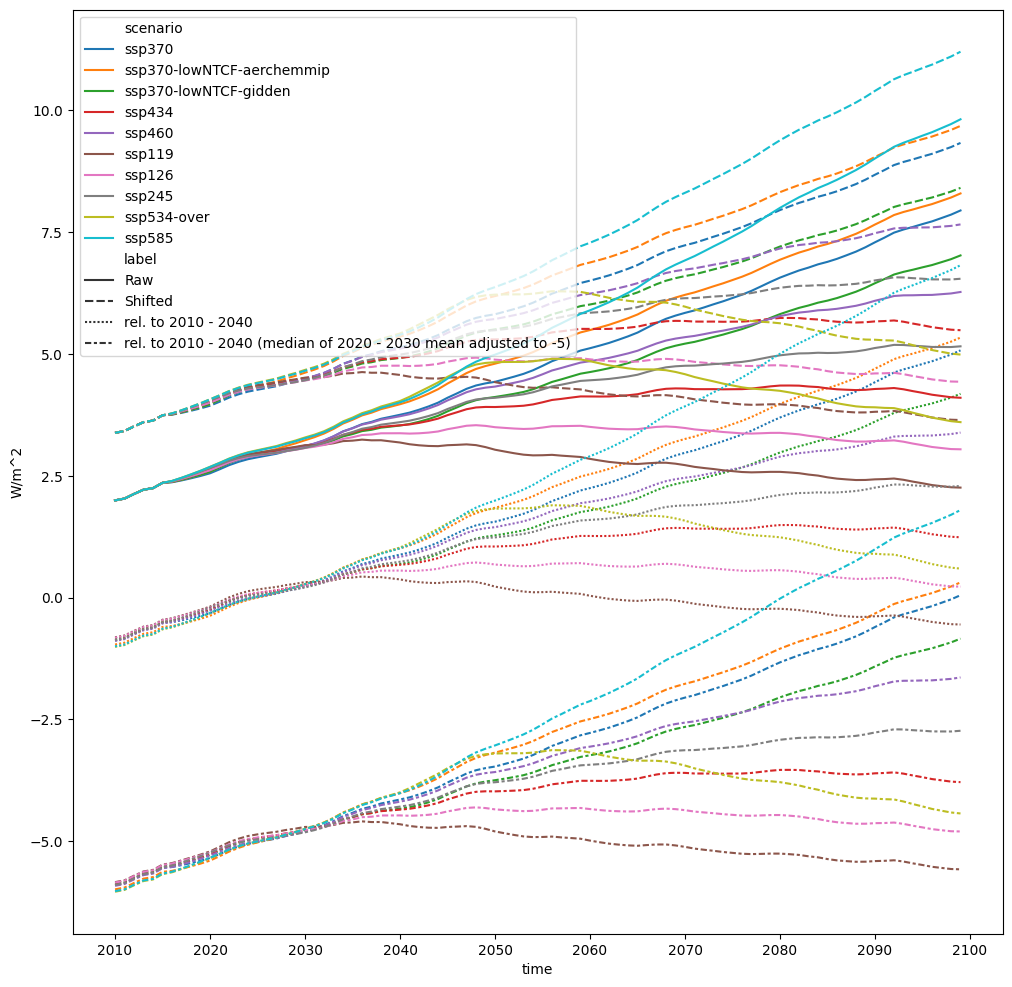
If we wanted, we could adjust the timeseries relative to some reference period first before doing the shift.
ref_period = range(2010, 2040 + 1)
erf_total_for_shift_rel_to_ref_period = erf_total_for_shift.relative_to_ref_period_mean(
year=ref_period
)
erf_total_for_shift_rel_to_ref_period[
"label"
] = f"rel. to {ref_period[0]} - {ref_period[-1]}"
target = -5
evaluation_period = range(2020, 2030 + 1)
erf_total_for_shift_rel_to_ref_period_shifted = (
erf_total_for_shift_rel_to_ref_period.adjust_median_to_target(
target=target,
evaluation_period=evaluation_period,
process_over=("scenario", "model"),
)
)
erf_total_for_shift_rel_to_ref_period_shifted[
"label"
] = "rel. to {} - {} (median of {} - {} mean adjusted to {})".format(
ref_period[0],
ref_period[-1],
evaluation_period[0],
evaluation_period[-1],
target,
)
pdf = run_append(
[
erf_total_for_shift,
erf_total_shifted,
erf_total_for_shift_rel_to_ref_period,
erf_total_for_shift_rel_to_ref_period_shifted,
]
)
fig, axes = plt.subplots(figsize=(12, 12))
pdf.lineplot(ax=axes, style="label")
<Axes: xlabel='time', ylabel='W/m^2'>